Biological Physics
Biological Physics aims to apply physical principles to living systems. It is an interdisciplinary science that combines biology with topics including statistical mechanics, thermodynamics, optics, spectroscopy, and data science. Training in biological physics leads to career opportunities in medical imaging and instrumentation, biotechnology, data science, and so much more.
The biological physics group at NC A&T uses experimental, theoretical and computational approaches to understand how the physical environment influences behavior and how organisms process physical signals to make sense of their surroundings.
Faculty
Dr. Chih Kuan Tung uses microfluidic devices to study how physical environments such as fluid flow and the mechanical properties of the fluid influence sperm motility. On the physics front, his research uses sperm collective swimming in viscoelastic fluid as a model system for polar active matter. On the biology front, he studies the role of mucus fluid viscoelasticity in sperm migration, including the collective swimming of sperm cells and swims against fluid flow.
Projects
A lot of predictions have been made for polar active matters either theoretically or numerically. In this project, we seek to examine them experimentally with our swimming sperm. This shines light on the natural noise structure within a biological system and its impact on statistical physics. Funded by NSF HRD 1665004.
Relevant Publications:
- Tung, C.-K., Lin, C., Harvey, B., Fiore, A. G., Ardon, F., Wu, M., & Suarez, S. S. (2017). Fluid viscoelasticity promotes collective swimming of sperm. Sci. Rep., 7, 3152. link
- Lin, C., Marks, T. K., Pajovic, M., Watanabe, S., & Tung, C.-k. (2018). Model parameter learning using Kullback–Leibler divergence. Physica A: Statistical Mechanics and its Applications, 491(Supplement C), 549-559. link
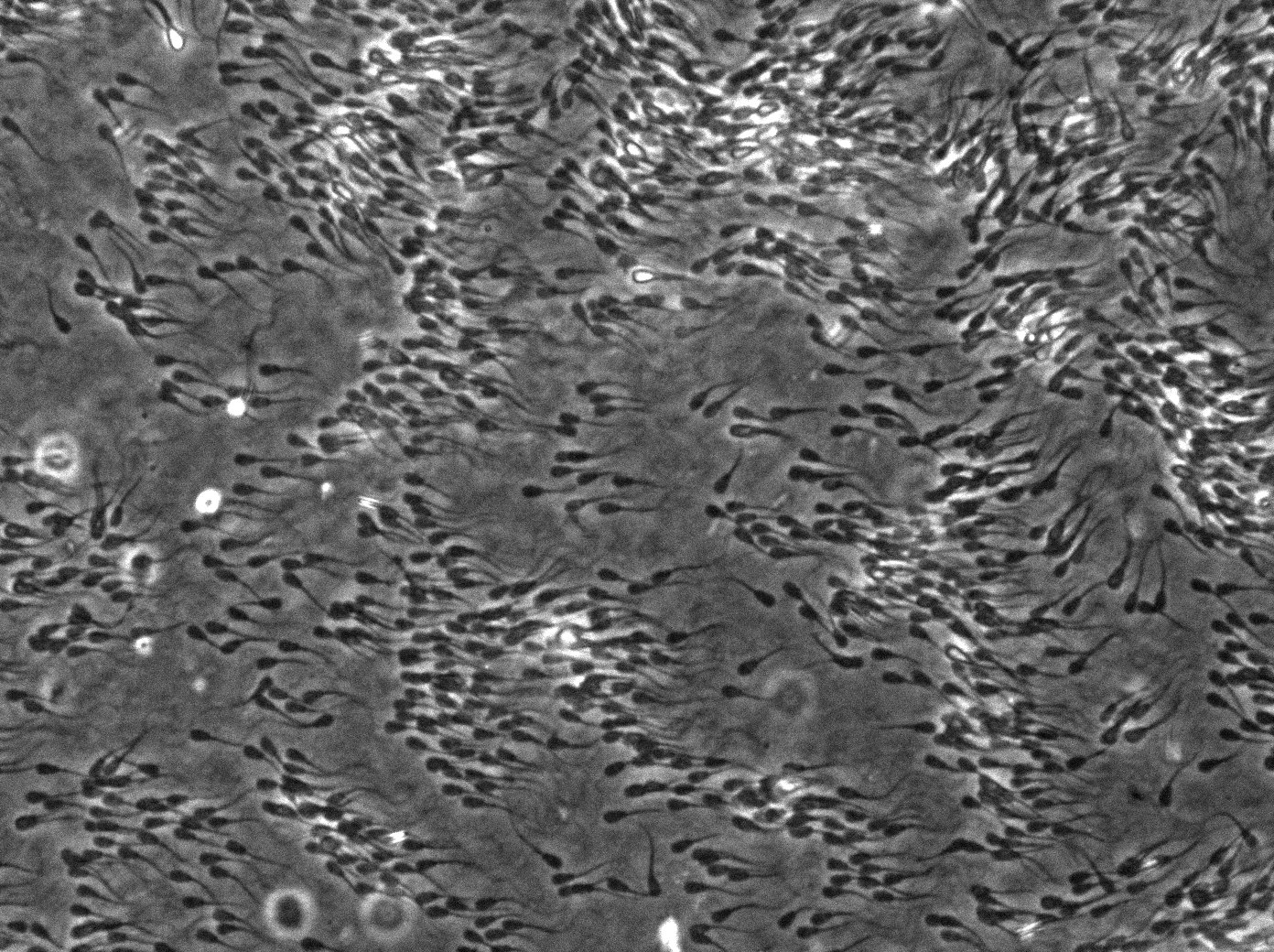 Here, we try to use a microfluidic device to model the liquid-liquid interface to see how sperm enter viscoelastic fluid and form collective swimming. Also, through imaging and analyses, we aim to find whether and how the collective swimming of sperm can be a biological advantage. This line of research advances our understanding in the context of evolutionary biology. Funded by NIH/NICHD R15HD095411.
Here, we try to use a microfluidic device to model the liquid-liquid interface to see how sperm enter viscoelastic fluid and form collective swimming. Also, through imaging and analyses, we aim to find whether and how the collective swimming of sperm can be a biological advantage. This line of research advances our understanding in the context of evolutionary biology. Funded by NIH/NICHD R15HD095411.
Relevant Publications:
- Tung, C.-K., Ardon, F., Fiore, A. G., Suarez, S. S., & Wu, M. (2014). Cooperative roles of biological flow and surface topography in guiding sperm migration revealed by a microfluidic model. Lab Chip, 14, 1348. link
-
Walker, B. J., Phuyal, S., Ishimoto, K., Tung, C.-K., & Gaffney, E. A. (2020). Computer-assisted beat-pattern analysis and the flagellar waveforms of bovine spermatozoa. Royal Society Open Science, 7(6), 200769. link
Opportunities
1. Human Psychophysics Experiments: We are looking for motivated undergraduate students interested in human vision and color perception to join our group to conduct color perception experiments. Students from Physics, Psychology, Computer Science, Engineering, and other related fields are welcome to apply. Some Matlab programing experience will be helpful. Contact vsingh@ncat.edu
2. Automated Cell Tracking: We are looking for motivated students from Physics, Computer Science, Engineering, and other related fields to join our group to develop computational tools to track bovine sperm cells. Students with good programming skills should contact send their CV to vsingh@ncat.edu or ctung@ncat.edu.
Contact ctung@ncat.edu or vsingh@ncat.edu to inquire about open positions.



